|
Biodiversity Information Science and Standards : Conference Abstract
|
|
Corresponding author: Urmas Kõljalg (urmas.koljalg@ut.ee)
Received: 17 Jun 2019 | Published: 26 Jun 2019
© 2019 Urmas Kõljalg, Kessy Abarenkov, R. Henrik Nilsson, Karl-Henrik Larsson, Andy F.S. Taylor
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Kõljalg U, Abarenkov K, Nilsson R, Larsson K, Taylor A (2019) The UNITE Database for Molecular Identification and for Communicating Fungal Species. Biodiversity Information Science and Standards 3: e37402. https://doi.org/10.3897/biss.3.37402
|
|
Abstract
UNITE (https://unite.ut.ee;
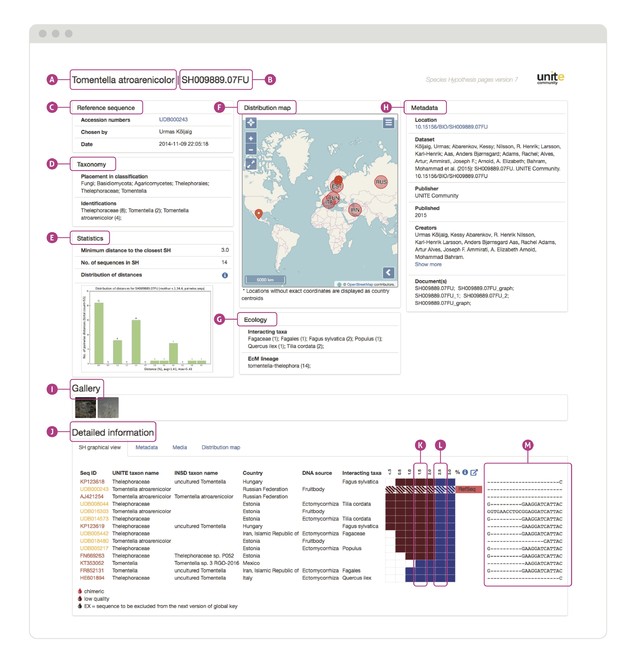
A screenshot of a UNITE SH Digital Object Identifier (DOI) page for Tomentella atroarenicolor (https://plutof.ut.ee/#/datacite/10.15156%2FBIO%2FSH009889.07FU). (A) The most accurate taxon name chosen automatically (or manually, if the default were overridden by an expert) from the available sequence identifications; (B) short ID of the DOI; (C) Data on reference sequence chosen to represent this SH; (D) placement of the SH in the fungal classification and identification records for individual sequences; the number after the taxon name indicates how many sequences carry that name. (E) Select statistics on the SH. The minimum distance 3.0% is the mandatory genetic difference between sister SHs. (F) Distribution map of the individual sequences. (G) Information on ecology (interacting taxa) if associated with the individual sequences. (H) DataCite-specific data on the DOI. (I) Images of the specimen or sample from which the DNA was extracted. Only a limited number of sequences have images attached to them. (J) Graphical overview of the SH with detailed information. (K) SH inclusiveness across sequence similarity threshold values. A threshold value (= minimum distance) of 1.5% will split these sequences into two SHs, shown here in different colours. (L) A threshold value of 2.5% will lump all sequences into a single SH. Each such SH is hyperlinked to its own unique web page. (M) Scrollable multiple sequence alignment of the SH. ‘RefSeq’ indicates that the sequence was selected manually to be the representative sequence for the SHs. RefSeqs stem from type specimens or other authentic and particularly trustworthy material. This particular SH contains both International Nucleotide Sequence Database Collaboration sequences (brown) and sequences that are only found in UNITE (yellow).
UNITE serves as a data provider for a range of metabarcoding software pipelines and regularly exchanges data with all major fungal sequence databases and other community resources.
Recent improvements include ITS-based species hypotheses for all eukaryotes and aggregation of full-length, high-quality ITS sequences generated by the PacBio Sequel system (https://www.pacb.com/products-and-services/sequel-system) from diverse material samples.
Keywords
molecular identification, persistent identifiers, fungi, taxonomy
Presenting author
Kessy Abarenkov
Presented at
Biodiversity_Next 2019
References
-
The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications.Nucleic Acids Research47:D259‑D264. https://doi.org/10.1093/nar/gky1022