|
Biodiversity Information Science and Standards : Conference Abstract
|
|
Corresponding author: Janno Harjes (janno7806@gmail.com)
Received: 21 Jun 2019 | Published: 02 Jul 2019
© 2019 Janno Harjes, Dagmar Triebel, Anton Link, Tanja Weibulat, Frank Oliver Glöckner, Gerhard Rambold
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Harjes J, Triebel D, Link A, Weibulat T, Glöckner F, Rambold G (2019) FAIR data in meta-omics research: Using the MOD-CO schema to describe structural and operational elements of workflows from field to publication. Biodiversity Information Science and Standards 3: e37596. https://doi.org/10.3897/biss.3.37596
|
|
Abstract
Nucleic acid and protein sequencing-based analyses are increasingly applied to determine origin, identity and traits of environmental (biological) objects and organisms. In this context, the need for corresponding data structures has become evident. As existing schemas and community standards in the domains of biodiversity and molecular biological research are comparatively limited with regard to the number of generic and specific elements, previous schemas for describing the physical and digital objects need to be replaced or expanded by new elements for covering the requirements from meta-omics techniques and operational details. On the one hand, schemas and standards are hitherto mostly focussed on elements, descriptors, or concepts that are relevant for data exchange and publication, on the other hand, detailed operational aspects regarding origin context and laboratory processing, as well as data management details, like the documentation of physical and digital object identifiers, are rather neglected.
The conceptual schema for Meta-omics Data and Collection Objects (MOD-CO; https://www.mod-co.net/) has been set up recently
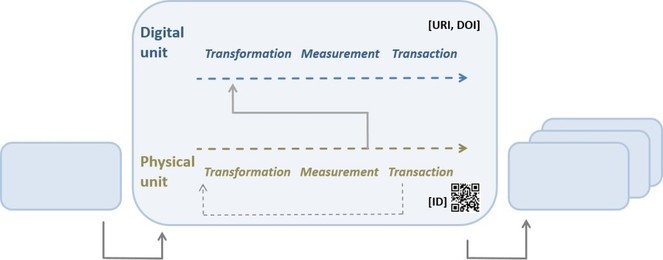
Workflow segments concatenated to a single workflow. A workflow segment comprises the elementary operations transformation, measurement and transaction being applied once or twice (due to preceding subsampling) to a physical object in focus, and the generation of data (measurement) and its subsequent transformation, measurement and transaction within the segment.
The MOD-CO concepts might be integrated as descriptor/element structures in the relational database DiversityDescriptions (DWB-DD) an open-source and freely available software of the Diversity Workbench (DWB; https://diversityworkbench.net/Portal/DiversityDescriptions; https://diversityworkbench.net). Currently, DWB-DD is installed at the Data Center of the Bavarian Natural History Collections (SNSB) to build an instance of its own for storing and maintaining MOD-CO-structured meta-omics research data packages and enrich them with ‘metadata’ elements from the community standards Ecological Markup Language (EML), Minimum Information about any (x) Sequence (MIxS), Darwin Core (DwC) and Access to Biological Collection Data (ABCD). These activities are achieved in the context of ongoing FAIR ('Findable, Accessible, Interoperable and Reuseable') biodiversity research data publishing via the German Federation for Biological Data (GFBio) network (https://www.gfbio.org/). Version 1.1 of the schema with extended collections of structural and operational design concepts is scheduled for 2020.
Keywords
collection data, conceptual schema, DiversityDescriptions, LIMS, meta-omics, workflow segments
Presenting author
Janno Harjes
Presented at
Biodiversity_Next 2019
Acknowledgements
We are grateful for discussions with Andreas Brachmann (Munich), Gregor Hagedorn (Berlin), Derek Peršoh (Bochum), Veronica Sanz (Munich), Carola Söhngen (Braunschweig), Thorsten Stoeck (Kaiserslautern), Christoph Tebbe (Braunschweig), and Pelin Yilmaz (Bremen).
References
-
Meta-omics data and collection objects (MOD-CO): a conceptual schema and data model for processing sample data in meta-omics research.Database2019https://doi.org/10.1093/database/baz002