|
Biodiversity Information Science and Standards : Conference Abstract
|
|
Corresponding author: Christian König (chr.koenig@outlook.com)
Received: 16 Jun 2019 | Published: 26 Jun 2019
© 2019 Christian König, Patrick Weigelt, Julian Schrader, Amanda Taylor, Jens Kattge, Holger Kreft
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: König C, Weigelt P, Schrader J, Taylor A, Kattge J, Kreft H (2019) Biodiversity Data Integration: The significance of data resolution and domain. Biodiversity Information Science and Standards 3: e37381. https://doi.org/10.3897/biss.3.37381
|
|
Abstract
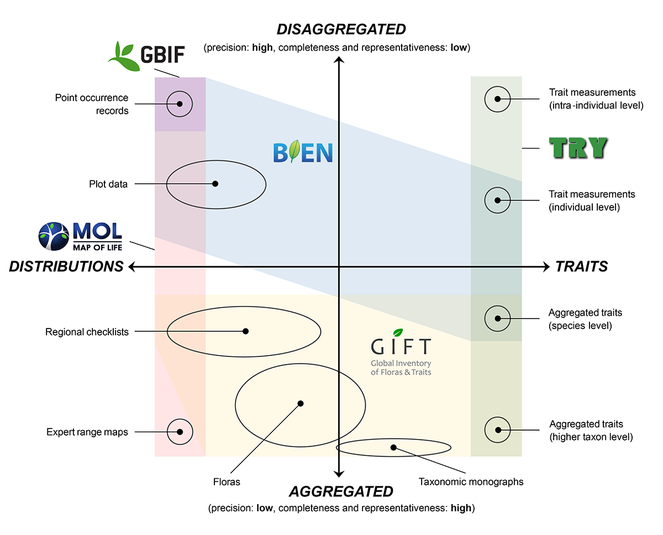
Recent years have seen an explosion in the availability of biodiversity data describing the distribution, function, and evolutionary history of life on earth. Integrating these heterogeneous data remains a challenge due to large variations in observational scales, collection purposes, and terminologies. Here, we conceptualize widely used biodiversity data types according to their domain (what aspect of biodiversity is described?) and informational resolution (how specific is the description?)(Fig.
Selected biodiversity data types, arranged according to their primary domain (here, species distributions versus functional traits) and informational resolution (disaggregated versus aggregated).
We use two case studies to substantiate our points. In the first case study, we show how coarse-grained regional plant checklists can be used to predict global growth form spectra at high spatial resolutions. Our predictions are highly consistent with fine-grained empirical data based on point occurrence records and vegetation plots. While such dis-aggregated data produce reliable results only for a limited set of well-covered regions, aggregated data types can provide critical information for the extrapolation of biodiversity patterns into less well-sampled regions. In the second case study, we revisit the latitudinal gradient in seed mass, which has been reported to exhibit a 320-fold decrease between 0 and 60 degrees latitude as well as a sharp, 7-fold drop at the edge of the tropics. Re-assessing this relationship using an independent dataset revealed a much more moderate, 11-fold decrease of seed mass towards the poles with little evidence for any abrupt changes. We show that the original results, which were based on relatively sparse, disaggregated data, are confounded by substantial biases in the representation of biomes and growth forms. This, we argue, amplified the magnitude and distorted the shape of the latitudinal gradient in seed mass.
In summary, this talk aims to provide both theoretical and practical arguments for a more integrated biodiversity data landscape, which utilizes the strengths and weaknesses of multiple data types across multiple domains and resolutions in order to produce more robust, comprehensive and informative macroecological inferences.
Keywords
data integration, functional biogeography, data resolution, representative, precision
Presenting author
Christian König
Presented at
Biodiversity_Next 2019