|
Biodiversity Information Science and Standards : Conference Abstract
|
|
Corresponding author: Lars Vogt (lars.m.vogt@googlemail.com)
Received: 12 Jun 2019 | Published: 19 Jun 2019
© 2019 Lars Vogt
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Vogt L (2019) Anatomy Knowledge Graphs: Toward FAIR morphological data. Biodiversity Information Science and Standards 3: e37203. https://doi.org/10.3897/biss.3.37203
|
|
Abstract
Most morphological data are still published as unstructured texts. This has far-reaching consequences for the Findability, Accessibility, Interoperability and Reusability of morphological data and thus for their FAIRness (
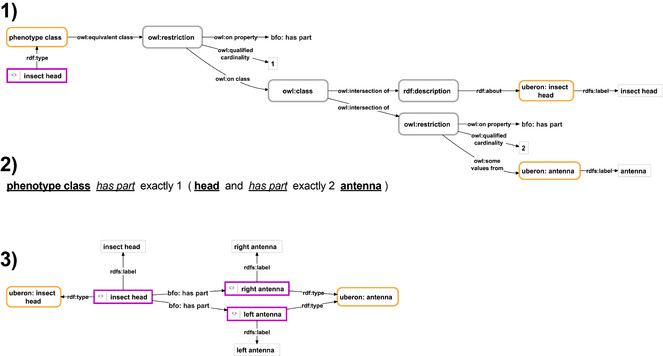
Phenotype description of an insect head with two antennae in form of a Semantic Phenotype in the form of an RDF-based graph or in Manchester syntax, and an Anatomy Knowledge Graph.
1) The Semantic Phenotype description of an insect head that has two antennae as its parts in the form of an RDF-based graph. It consists of an instance (purple-bordered box) that instantiates the phenotype class that contains the actual description of the phenotype (yellow-bordered box) in form of a class axiom consisting of anonymous property restrictions and class descriptions (grey-bordered boxes). The class axiom characterizes all instances of the class to consist of exactly one instance of insect head (id UBERON:6000004) that has as its parts exactly two instances of antenna (id UBERON:0000972). 2) An alternative format of the same Semantic Phenotype representing the class axioms from the ‘phenotype class’ from (1) expressed in OWL Manchester Syntax (ontology classes shown with their label in bold and underlined, ontology properties with their label in italics and underlined, ‘and’ being used in the sense of intersection of two mathematical sets and ‘exactly’ as a cardinality specification). 3) The Anatomy Knowledge Graph description of an insect head that has two antennae as its parts. It consists of the instance of insect head (id UBERON:6000004) and two instances of antenna (id UBERON:0000972), which relate to the head as its parts. Labels (in light-grey-bordered boxes) indicate how the different instances should be represented in a human-readable format. For reasons of clarity, resources are not represented with their URIs but with labels. purple-bordered box = ontology instance; yellow-bordered box with rounded corners = ontology class; grey-bordered box with rounded corners = anonymous class; light-grey-bordered box = literal or numerical value; labeled arrow = property.
Keywords
Anatomy Knowledge Graph, semantic phenotype, FAIR data principle, anatomy, morphological description, semantic graph, knowledge graph, ontology
Presenting author
Lars Vogt
Presented at
Biodiversity_Next 2019
Acknowledgements
I thank Peter Grobe, Roman Baum, Christian Köhler, Sandra Meid, Björn Quast, Stefan Richter, Christian Wirkner, Michael Ohl, Stefan Graf, Jan Decher, and Christian Montermann for discussing the here presented ideas. I thank István Mikó, Matt Yoder, and Torben Göpel for discussing some of the data schemes for representing morphological structures.
Funding program
VO 1244/8-1
Grant title
eScience-Compliant Standards for Morphology
Hosting institution
Rheinische Friedrich-Wilhelms-Universität, Bonn, Germany
References
-
The FAIR Guiding Principles for scientific data management and stewardship.Scientific Data3:160018. https://doi.org/10.1038/sdata.2016.18