|
Biodiversity Information Science and Standards : Conference Abstract
|
|
Corresponding author: Mikko Heikkinen (mikko.heikkinen@helsinki.fi)
Received: 12 Jun 2019 | Published: 18 Jun 2019
© 2019 Mikko Heikkinen, Ville-Matti Riihikoski, Anniina Kuusijärvi, Dare Talvitie, Tapani Lahti, Leif Schulman
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Heikkinen M, Riihikoski V, Kuusijärvi A, Talvitie D, Lahti T, Schulman L (2019) Kotka - A national multi-purpose collection management system. Biodiversity Information Science and Standards 3: e37179. https://doi.org/10.3897/biss.3.37179
|
|
Abstract
Many natural history museums share a common problem: a multitude of legacy collection management systems (CMS) and the difficulty of finding a new system to replace them. Kotka is a CMS created by the Finnish Museum of Natural History (Luomus) to solve this problem. Its development started in late 2011 and was put into operational use in 2012. Kotka was first built to replace dozens of in-house systems previously used at Luomus, but eventually grew into a national system, which is now used by 10 institutions in Finland. Kotka currently holds c. 1.7 million specimens from zoological, botanical, paleontological, microbial and botanic garden collections, as well as data from genomic resource collections. Kotka is designed to fit the needs of different types of collections and can be further adapted when new needs arise.
Kotka differs in many ways from traditional CMS's. It applies simple and pragmatic approaches. This has helped it to grow into a widely used system despite limited development resources – on average less than one full-time equivalent developer (FTE).
The aim of Kotka is to improve collection management efficiency by providing practical tools. It emphasizes the quantity of digitized specimens over completeness of the data. It also harmonizes collection management practices by bringing all types of collections under one system.
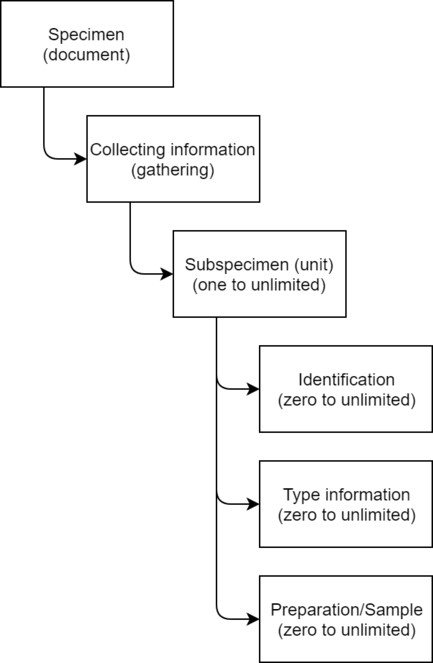
Kotka stores data mostly in a denormalized free text format using a triplestore and a simple hierarchical data model (Fig.
Specimen data model used by Kotka. Every specimen can have any number of sub-specimens (e.g. insects in a jar), and each of them any number of identifications, type information and preparations (morphological preparations, DNA extracts, tissue samples or such).
Kotka is a web application and data can be entered, edited, searched and exported through a browser-based user interface. However, most users prefer to enter new data in customizable MS-Excel templates, which support the hierarchical data model, and upload these to Kotka. Batch updates can also be done using Excel. Kotka stores all revisions of the data to avoid any data loss due to technical or human error.
Kotka also supports designing and printing specimen labels, annotations by external users, as well as handling accessions, loan transactions, and the Nagoya protocol. Taxonomy management is done using a separate system provided by the Finnish Biodiversity Information Facility (FinBIF). This decoupling also allows entering specimen data before the taxonomy is updated, which speeds up specimen digitization. Every specimen is given a persistent unique HTTP-URI identifier (CETAF stable identifiers). Specimen data is accessible through the FinBIF portal at species.fi, and will later be shared to GBIF according to agreements with data holders.
Kotka is continuously developed and adapted to new requirements in close collaboration with curators and technical collection staff, using agile software development methods. It is available as open source, but is tightly integrated with other FinBIF infrastructure, and currently only offered as an online service (Software as a Service) hosted by FinBIF.
Keywords
collection management system, natural history museum collection, denormalized database, web application
Presenting author
Mikko Heikkinen
Presented at
Biodiversity_Next 2019