|
Biodiversity Information Science and Standards : Conference Abstract
|
|
Corresponding author: Abraham Nieva de la Hidalga (nievadelahidalgaa@cardiff.ac.uk)
Received: 11 Jun 2019 | Published: 18 Jun 2019
© 2019 Abraham Nieva de la Hidalga, David Owen, Irena Spasic, Paul Rosin, Xianfang Sun
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Nieva de la Hidalga A, Owen D, Spacic I, Rosin P, Sun X (2019) Use of Semantic Segmentation for Increasing the Throughput of Digitisation Workflows for Natural History Collections. Biodiversity Information Science and Standards 3: e37161. https://doi.org/10.3897/biss.3.37161
|
|
Abstract
The need to increase global accessibility to specimens while preserving the physical specimens by reducing their handling motivates digitisation. Digitisation of natural history collections has evolved from recording of specimens’ catalogue data to including digital images and 3D models of specimens. The sheer size of the collections requires developing high throughput digitisation workflows, as well as novel acquisition systems, image standardisation, curation, preservation, and publishing. For instance, herbarium sheet digitisation workflows (and fast digitisation stations) can digitise up to 6,000 specimens per day; operating digitisation stations in parallel can increase that capacity. However, other activities of digitisation workflows still rely on manual processes which throttle the speed with which images can be published. Image quality control and information extraction from images can benefit from greater automation.
This presentation explores the advantages of applying semantic segmentation (Fig.
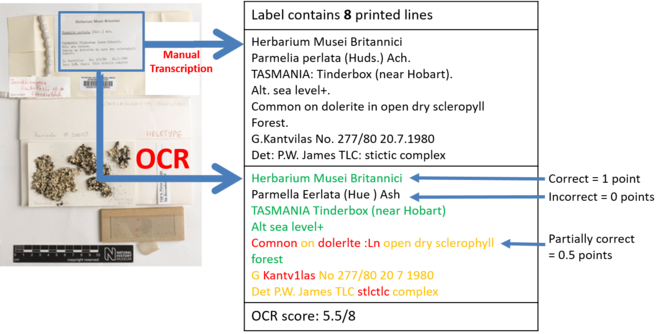
Example of semantic segmentation of a herbarium sheet. Semantic segmentation entails the identification and classification of image elements. Four classes are targeted for identification use in: labels, barcodes, clour charts and scale. IQM uses labels and barcodes, while IEFI experiments targeted labels and barcodes.
The IQM experiments evaluated the application of three quality attributes to full images and to image segments: colourfulness (Fig.
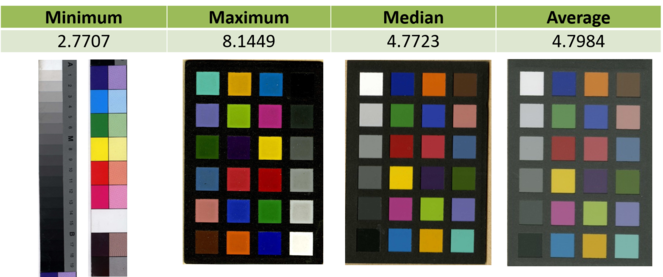
Comparison of results of calculating colurfulness on segmented colour charts from herbarium sheets set (Algorithm from
Comparison of results of calculating contrast on segmented colour charts from herbarium sheets set (Algorithm from
Comparison of results of calculating sharpness on barcode segments from herbarium sheets set (Algorithm from
The IEFI experiments compared the performance of four optical character recognition (OCR) programs applied to full images (
Comparison processing times of three OCR programs applied to full images (250 images) and to individual segments (1,837 segments from those images).
|
|
250 whole images |
1,837 segments |
Difference (h:m:s) |
|
Tesseract 4.x (Tess 4J) |
01:06:05 |
00:45:02 |
-00:21:03 |
|
Tesseract 3.x (Tess 4J) |
00:50:02 |
00:23:17 |
-00:26:45 |
|
Abbyy FineReader Engine 12 |
01:18:15 |
00:29:24 |
-00:48:51 |
|
Processing Time (h:m:s) |
|||
Comparison of line correctness for four OCR programs applied to full images (5 images) and to individual segments (22 segments from those images).
|
|
5 whole images Mean line correctness (%) |
22 segments Mean line correctness (%) |
Difference (%) |
|
Tesseract 4.x |
72.8 |
75.2 |
+2.4% |
|
Tesseract 3.x |
44.1 |
63.7 |
+19.6% |
|
Abbyy FineReader Engine 12 |
61.0 |
77.3 |
+16.3% |
|
Microsoft OneNote 2013 |
78.9 |
65.5 |
-13.4% |
Line correctness evaluation example. Line correctness means that information from lines is ordered, not fragmented, and not mixed.
The findings support the feasibility of further automation of digitisation workflows for natural history collections. In addition to increasing the accuracy and speed of IQM and IEFI activities, the explored approaches can be packaged and published, enabling automated quality management and information extraction to be offered as a service, taking advantage of cloud platforms and workflow engines.
Keywords
semantic segmentation, image quality management, optical character recognition, digitisation, natural history collections, digital specimens
Presenting author
Abraham Nieva de la Hidalga
Presented at
Biodiversity_Next 2019
Funding program
Horizon 2020 Framework Programme of the European Union
Grant title
ICEDIG – “Innovation and consolidation for large scale digitisation of natural heritage” H2020-INFRADEV-2016-2017 – Grant Agreement No. 777483
References
-
A fast approach for no-reference image sharpness assessment based on maximum local variation.IEEE Signal Processing Letters21(6):751‑755. https://doi.org/10.1109/LSP.2014.2314487
-
A benchmark dataset of herbarium specimen images with label data.Biodiversity Data Journal7https://doi.org/10.3897/bdj.7.e31817
-
The use of Optical Character Recognition (OCR) in the digitisation of herbarium specimen labels.PhytoKeys38:15‑30. https://doi.org/10.3897/phytokeys.38.7168
-
Semi-supervised semantic and instance segmentation. https://github.com/NaturalHistoryMuseum/semantic-segmentation. Accessed on: 2018-4-26.
-
Measuring colorfulness in natural images.Human Vision and Electronic Imaging VIIIhttps://doi.org/10.1117/12.477378
-
Global Contrast Factor-a new approach to Image contrast.Computational Aesthetics159‑168.
-
Colorfulness of the image: definition, computation, and properties.Conference on Lightmetry and Light and Optics in Biomedicine709https://doi.org/10.1117/12.675760
-
The visual dictionary of photography.AVA Publishing[ISBN978-2-940411-04-7] https://doi.org/10.5040/9781350088733