|
Biodiversity Information Science and Standards : Conference Abstract
|
|
Corresponding author: Dmitry S. Schigel (dschigel@gbif.org)
Received: 10 Jun 2019 | Published: 13 Jun 2019
This is an open access article distributed under the terms of the CC0 Public Domain Dedication.
Citation: Schigel D, Jeppesen T, Finn R, Cochrane G, Kõljalg U, Quast C, Lanfear J, Orrell T, Hobern D, Miller J (2019) Going Molecular: Sequence-based spatiotemporal biodiversity evidence in GBIF. Biodiversity Information Science and Standards 3: e37036. https://doi.org/10.3897/biss.3.37036
|
|
Abstract
The Global Biodiversity Information Facility (GBIF) was established by governments in 2001, largely through the initiative and leadership of the natural history collections community, following the 1999 recommendation by a working group under the Megascience Forum (predecessor of the Global Science Forum) of the Organization for Economic Cooperation and Development (OECD). Over 20 years, GBIF has helped develop standards and convened a global community of data-publishing institutions, aggregrating over one billion specimen occurrence records freely and openly available for use in research and policy making. These GBIF mediated data range from vouchered museum specimens to observation records generated by humans and machines. New data are being generated from integrated remote sensing, ecological sampling, and molecular sequencing that have strong geospatial components but lack traditional vouchers. GBIF is working with partners to develop best practices of bringing this data into the GBIF architecture.
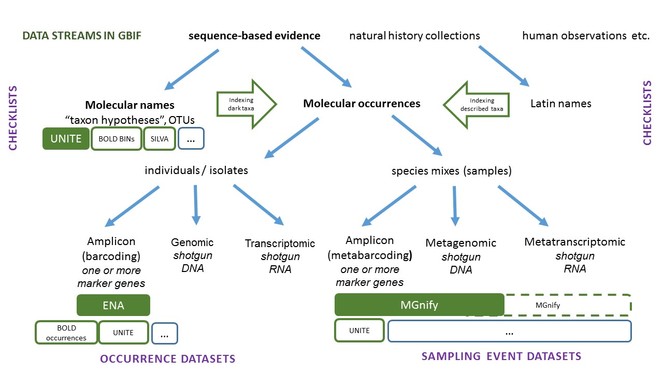
Following discussions during the second Global Biodiversity Information Conference in 2018, GBIF and the European Bioinformatics Institute (EMBL-EBI), supported by ELIXIR, have extended collaboration to share species occurrence records known only from their genetic material. When these data providers contribute data coordinates along with the sequences to the European Nucleotide Archive (ENA), the records will appear on GBIF maps and in spatial searches. This collaboration enables significant new molecular data streams to become discoverable through GBIF.org: by mid-March 2019, over 7.8m individual occurrence records via the ENA, and over 13.2m records as standardized Darwin Core sampling-event datasets via MGnify, a resource that provides taxonomic and functional annotations on sequences derived from environmental sequencing projects. Sequence-based occurrence records published by ENA and MGnify boost representation of microbial diversity which was underrepresented at GBIF. The ELIXIR-ENA-MGnify-GBIF partnership is working on further refinement of the dynamic data linkages, frequency of updates and other improvements. The API-based tool that connects GBIF data infrastructures is open to new data contributors and for indexes of molecular occurrences. Indexing of these data streams is dependent on the presence of a name (any rank) with the sequence.
Under the current Codes of nomenclature, animals, fungi, plants, and algae cannot be described based on exclusively sequence data. Yet, a significant volume of biodiversity data has only been represented by DNA sequences. Barcoding and sequence clustering procedures vary among taxa and research communities, but clusters can be related to a taxon with a Latin name. Many DNA similarity clusters do not contain a sequence from a formally described taxon; however these sequence clusters provide provisional molecular names for nomenclatural communication. In the best cases, curated libraries of reference sequences, their metadata, clusters, alignments, and links to individuals and physical material become de facto naming conventions for certain taxonomic groups, and co-exist with Latin names.
Integration of molecular names into the taxonomic backbone of GBIF started with Fungi and UNITE, a data management and identification environment for fungal ITS barcodes with 87,000+ fungal species hypotheses demarcating 800,000+ sequence specimens as of March 2019. Checklist publication of all names in UNITE through GBIF.org including Linnaean names and stable, DOI-trackable molecular sequence based ‘species hypotheses’, enables indexing of fungal metabarcoding data worldwide, such as BIOWIDE. As names are currently essential to indexing the world’s occurrence data, GBIF will develop similar linkages with names in the Barcode of Life data system (BOLD) and in SILVA - a resource for high-quality ribosomal RNA sequence data and taxonomy, and welcomes other reference systems to this development. Expanding the molecular data streams (Fig.
Presenting author
Dmitry S. Schigel
Presented at
Biodiversity_Next 2019