|
Biodiversity Information Science and Standards : Conference Abstract
|
|
Corresponding author: Mariya Dimitrova (m.dimitrova@pensoft.net)
Received: 02 Apr 2019 | Published: 18 Jun 2019
© 2019 Mariya Dimitrova, Viktor Senderov, Teodor Georgiev, Georgi Zhelezov, Lyubomir Penev
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Dimitrova M, Senderov V, Georgiev T, Zhelezov G, Penev L (2019) OpenBiodiv: Linking Type Materials, Institutions, Locations and Taxonomic Names Extracted From Scholarly Literature. Biodiversity Information Science and Standards 3: e35089. https://doi.org/10.3897/biss.3.35089
|
|
Abstract
OpenBiodiv is a knowledge management system containing biodiversity knowledge extracted from scholarly literature: both recently published articles in Pensoft's journals and legacy (taxon treatments extracted by Plazi) (
The OpenBiodiv workflow: from information extraction to answering complex biodiversity questions.
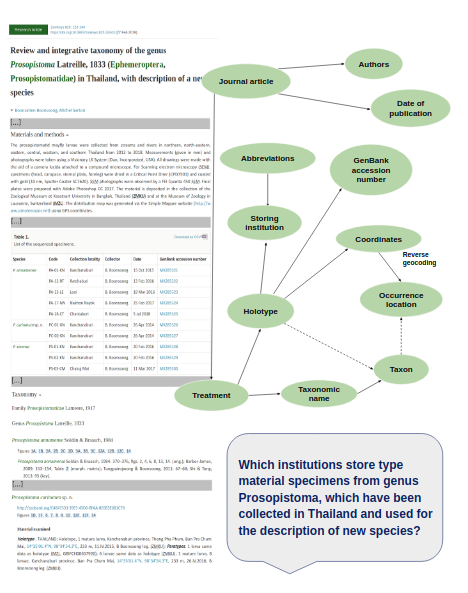
The ontology allows to model the structure of research articles and treatments, as well as their corresponding metadata. Thus, OpenBiodiv-O is used to represent not only the sections of treatments but also the various entities within them, for instance geographic coordinates and institution codes within the “Type materials” section of a treatment. Institution codes marked up within articles using the Darwin Core standard (
Keywords
Biodiversity Knowledge Graph, Semantic Technologies, Use Case, Ontology, Information Extraction
Presenting author
Mariya Dimitrova
References
-
Taxonomic information exchange and copyright: the Plazi approach.BMC Research Notes2(1):53‑53. https://doi.org/10.1186/1756-0500-2-53
-
XML schemas and mark-up practices of taxonomic literature.ZooKeys150:89‑116. https://doi.org/10.3897/zookeys.150.2213
-
OpenBiodiv: an Implementaion of a Semantic System Running on top of the Biodiversity Knowledge Graph.Biodiversity Information Science and Standards1:e20084‑e20084. https://doi.org/10.3897/tdwgproceedings.1.20084
-
OpenBiodiv-O: ontology of the OpenBiodiv knowledge management system.Journal of Biomedical Semantics9(1). https://doi.org/10.1186/s13326-017-0174-5
-
Darwin Core: An Evolving Community-Developed Biodiversity Data Standard.PLOS ONE7(1):e29715‑e29715. https://doi.org/10.1371/journal.pone.0029715