|
Biodiversity Information Science and Standards : Conference Abstract
|
|
Corresponding author: Franck Michel (franck.michel@cnrs.fr)
Received: 14 Apr 2018 | Published: 22 May 2018
© 2018 Franck Michel, The Bioschemas Community
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Michel F, The Bioschemas Community (2018) Bioschemas & Schema.org: a Lightweight Semantic Layer for Life Sciences Websites. Biodiversity Information Science and Standards 2: e25836. https://doi.org/10.3897/biss.2.25836
|
|
Abstract
Web portals are commonly used to expose and share scientific data. They enable end users to find, organize and obtain data relevant to their interests. With the continuous growth of data across all science domains, researchers commonly find themselves overwhelmed as finding, retrieving and making sense of data becomes increasingly difficult. Search engines can help find relevant websites, but the short summarizations they provide in results lists are often little informative on how relevant a website is with respect to research interests.
To yield better results, a strategy adopted by Google, Yahoo, Yandex and Bing involves consuming structured content that they extract from websites. Towards this end, the schema.org collaborative community defines vocabularies covering common entities and relationships (e.g., events, organizations, creative works) (
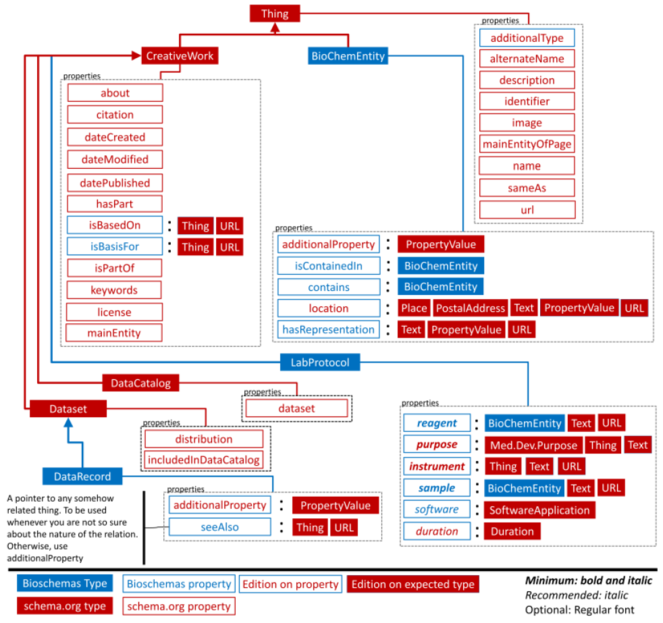
Although schema.org encompasses terms related to data repositories, datasets, citations, events, etc., it lacks specialized terms for modeling research entities. The Bioschemas community (
The success of schema.org lies in its simplicity and the support by major search engines. By extending schema.org, Bioschemas enables life sciences research communities to benefit from a lightweight semantic layer on websites and thus facilitates discoverability and interoperability across them. From an initial pilot including just a few bio-types such as proteins and samples, the Bioschemas community has grown and is now opening up towards other disciplines. The biodiversity domain is a promising candidate for such further extensions. We can think of additional profiles to account for biodiversity-related information. For instance, since taxonomic registers are the backbone of many web portals and databases, new profiles could describe taxa and scientific names while reusing well-adopted vocabularies such as Darwin Core terms (
Presenting author
Franck Michel
References
-
Lessons Learned from Adapting the Darwin Core Vocabulary Standard for Use in RDF.Semantic Web7(6):617‑627.
-
Bioschemas: schema.org for the Life Sciences.Proceedings of SWAT4LS.CEUR,2042
-
Schema.org: Evolution of Structured Data on the Web.Communications of the ACM59(2):44‑51.
-
Report of the TDWG Vocabulary Management Task Group (VoMaG). http://www.gbif.org/document/80862