|
Biodiversity Information Science and Standards : Conference Abstract
|
|
Corresponding author: Teodor Georgiev (preprint@pensoft.net)
Received: 12 Apr 2018 | Published: 17 May 2018
© 2018 Lyubomir Penev, Donat Agosti, Teodor Georgiev, Viktor Senderov, Guido Sautter, Terry Catapano, Pavel Stoev
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Penev L, Agosti D, Georgiev T, Senderov V, Sautter G, Catapano T, Stoev P (2018) The Open Biodiversity Knowledge Management (eco-)System: Tools and Services for Extraction, Mobilization, Handling and Re-use of Data from the Published Literature. Biodiversity Information Science and Standards 2: e25748. https://doi.org/10.3897/biss.2.25748
|
|
Abstract
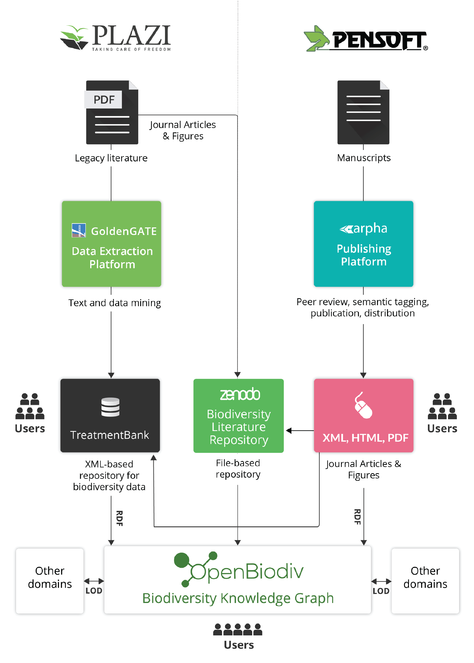
The Open Biodiversity Knowledge Management System (OBKMS) is an end-to-end, eXtensible Markup Language (XML)- and Linked Open Data (LOD)-based ecosystem of tools and services that encompasses the entire process of authoring, submission, review, publication, dissemination, and archiving of biodiversity literature, as well as the text mining of published biodiversity literature (Fig.
OBKMS is the result of a joint endeavour by Plazi and Pensoft lasting many years. The system was developed with the support of several biodiversity informatics projects - initially (Virtual Biodiversity Research and Access Network for Taxonomy) ViBRANT, and then followed by pro-iBiosphere, European Biodiversity Observation Network (EU BON), and Biosystematics, informatics and genomics of the big 4 insect groups (BIG4). The system includes the following key components:
-
ARPHA Journal Publishing Platform: a journal publishing platform based on the TaxPub XML extension for National Library of Medicine (NLM)’s Journal Publishing Document Type Definition (DTD) (Version 3.0). Its advanced ARPHA-BioDiv component deals with integrated biodiversity data and narrative publishing (
Penev et al. 2017 ). -
GoldenGATE Imagine: an environment for marking up, enhancing, and extracting text and data from PDF files, supporting the TaxonX XML schema. It has specific enhancements for articles containing descriptions of taxa ("taxonomic treatments") in the field of biological systematics, but its core features may be used for general purposes as well.
-
Biodiversity Literature repository (BLR): a public repository hosted at Zenodo (CERN) for published articles (PDF and XML) and images extracted from articles.
-
Ocellus/Zenodeo: a search interface for the images stored at BLR.
-
TreatmentBank: an XML-based repository for taxonomic treatments and data therein extracted from literature.
-
The OpenBiodiv knowledge graph: a biodiversity knowledge graph built according to the Linked Open Data (LOD) principles. Uses the RDF data model, the SPARQL Protocol and RDF Query Language (SPARQL) query language, is open to the public, and is powered by the OpenBiodiv-O ontology (
Senderov et al. 2018 ). -
-
Semantic search and browser for the biodiversity knowledge graph.
-
Multiple semantic apps packaging specific views of the biodiviersity knowledge graph.
-
-
Supporting tools:
-
Pensoft Markup Tool (PMT)
-
ARPHA Writing Tool (AWT)
-
ReFindit
-
R libraries for working with RDF and for converting XML to RDF (ropenbio, RDF4R).
-
Plazi RDF converter, web services and APIs.
-
As part of OBKMS, Plazi and Pensoft offer the following services beyond supplying the software toolkit:
-
Digitization through imaging and text capture of paper-based or digitally born (PDF) legacy literature.
-
XML markup of both legacy and newly published literature (journals and books).
-
Data extraction and markup of taxonomic names, literature references, taxonomic treatments and organism occurrence records.
-
Export and storage of text, images, and structured data in data repositories.
-
Linking and semantic enhancement of text and data, bibliographic references, taxonomic treatments, illustrations, organism occurrences and organism traits.
-
Re-packaging of extracted information into new, user-demanded outputs via semantic apps at the OpenBiodiv portal.
-
Re-publishing of legacy literature (e.g., Flora, Fauna, and Mycota series, important biodiversity monographs, etc.).
-
Semantic open access publishing (including data publishing) of journal and books.
-
Integration of biodiversity information from legacy and newly published literature into interoperable biodiversity repositories and platforms (Global Biodiversity Information Facility (GBIF), Encyclopedia of Life (EOL), Species-ID, Plazi, Wikidata, and others).
In this presentation we make the case for why OpenBiodiv is an essential tool for advancing biodiversity science. Our argument is that through OpenBiodiv, biodiversity science makes a step towards the ideals of open science (
A particular example of how OpenBiodiv can advance biodiversity science is demonstrated by the OpenBiodiv's solution to "taxonomic anarchy" (
Presenting author
Teodor Georgiev
References
-
Names are not good enough: Reasoning over taxonomic change in the Andropogon complex1.Semantic Web7(6):645‑667. https://doi.org/10.3233/sw-160220
-
Perspectives: Towards a language for mapping relationships among taxonomic concepts.Systematics and Biodiversity7(1):5‑20. https://doi.org/10.1017/s147720000800282x
-
Taxonomy anarchy hampers conservation.Nature546(7656):25‑27. https://doi.org/10.1038/546025a
-
ARPHA-BioDiv: A Toolbox for Scholarly Publication and Dissemination of Biodiversity Data Based on the ARPHA Publishing Platform.Research Ideas and Outcomes3:e13088. https://doi.org/10.3897/rio.3.e13088
-
The Open Biodiversity Knowledge Management System in Scholarly Publishing.Research Ideas and Outcomes2:e7757. https://doi.org/10.3897/rio.2.e7757
-
OpenBiodiv-O: ontology of the OpenBiodiv knowledge management system.Journal of Biomedical Semantics9(1):5. https://doi.org/10.1186/s13326-017-0174-5