|
Proceedings of TDWG : Conference Abstract
|
|
Corresponding author: Yue Zhu (yue.zhu@polytechnique.edu), Régine Vignes-Lebbe (regine.vignes_lebbe@upmc.fr)
Received: 17 Aug 2017 | Published: 18 Aug 2017
© 2017 Yue Zhu, Thibaut Durand, Eric Chenin, Marc Pignal, Patrick Gallinari, Régine Vignes-Lebbe
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Zhu Y, Durand T, Chenin E, Pignal M, Gallinari P, Vignes-Lebbe R (2017) Using a Deep Convolutional Neural Network for Extracting Morphological Traits from Herbarium Images. Proceedings of TDWG 1: e20400. https://doi.org/10.3897/tdwgproceedings.1.20400
|
|
Abstract
Natural history collection data are now accessible through databases and web portals. However, ecological or morphological traits describing specimens are rarely recorded while gathering data. This lack limits queries and analyses. Manual tagging of millions of specimens will be a huge task even with the help of citizen science projects such as “les herbonautes” (http://lesherbonautes.mnhn.fr). On the other hand, deep learning methods that use convolutional neural networks (CNN) demonstrate their efficiency in various domains, such as computer vision (
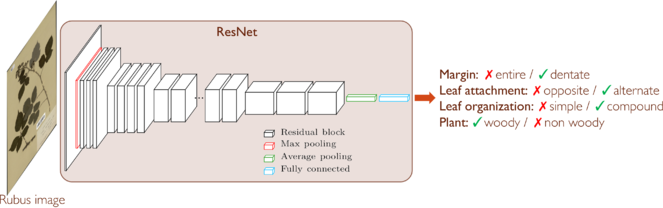
We aim to use deep learning to provide a visual representation of words used to describe plants (e.g. simple, or compound leaf), and to associate those words with specimens in the Paris herbarium. This will provide a semantic description of each of the 7 millions images of the fully digitized collection of the Paris herbarium in the Muséum National d'Histoire Naturelle (MNHN, Paris, France). In a proof of concept project, we have used a CNN - pre-trained on the image database ImageNet (http://www.image-net.org) - in order to identify 4 morphological traits of leaves, using 103,000 herbarium images from 11 different taxa: margin (entire / dentate), leaf attachment (opposite / alternate), leaf organization (simple / compound), plant (woody / non-woody) (see Fig.
Keywords
Deep learning, convolutionnal neural network, herbarium, image, morphology
Presenting author
Régine Vignes-Lebbe
References
-
Convolutional Neural Networks for Speech Recognition.IEEE/ACM Transactions on Audio, Speech, and Language Processing22(10):1533‑1545. https://doi.org/10.1109/taslp.2014.2339736
-
Factors of Transferability for a Generic ConvNet Representation.IEEE Transactions on Pattern Analysis and Machine Intelligence38(9):1790‑1802. https://doi.org/10.1109/tpami.2015.2500224
-
WILDCAT: Weakly Supervised Learning of Deep ConvNets for Image Classification, Pointwise Localization and Segmentation.29th IEEE Conference on Computer Vision and Pattern Recognition.
-
Chimpanzee faces in the wild: Log-Euclidean CNNs for predicting identities and attributes of primates. In: Rosenhahn B, Bjoern A (Eds)Pattern recognition 38th German conference, GCPR 2016.Springer International Publishing AG,Hannover, Germany
-
ImageNet Classification with Deep Convolutional Neural Networks.NIPS 2012.
-
IEEE Conference on Computer Vision and Pattern Recognition (CVPR).2015. https://doi.org/10.1109/cvpr.2015.7299170