|
Proceedings of TDWG : Conference Abstract
|
|
Corresponding author: Viktor Senderov (datascience@pensoft.net)
Received: 11 Aug 2017 | Published: 11 Aug 2017
© 2017 Viktor Senderov, Teodor Georgiev, Donat Agosti, Terry Catapano, Guido Sautter, Éamonn Ó Tuama, Nico Franz, Kiril Simov, Pavel Stoev, Lyubomir Penev
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Senderov V, Georgiev T, Agosti D, Catapano T, Sautter G, Ó Tuama É, Franz N, Simov K, Stoev P, Penev L (2017) OpenBiodiv Computer Demo: an Implementation of a Semantic System Running on top of the Biodiversity Knowledge Graph. Proceedings of TDWG 1: e20193. https://doi.org/10.3897/tdwgproceedings.1.20193
|
|
Abstract
We present OpenBiodiv - an implementation of the Open Biodiversity Knowledge Management System. We believe OpenBiodiv is possibly the first pilot-stage implenatation of a semantic system running on top of the biodiversity knowledge graph.
The need for an integrated information system serving the needs of the biodiversity community can be dated at least as far back as the sanctioning of the Bouchout declaration in 2007. The Bouchout declaration proposes to make biodiversity knowledge freely available as Linked Open Data (LOD)*
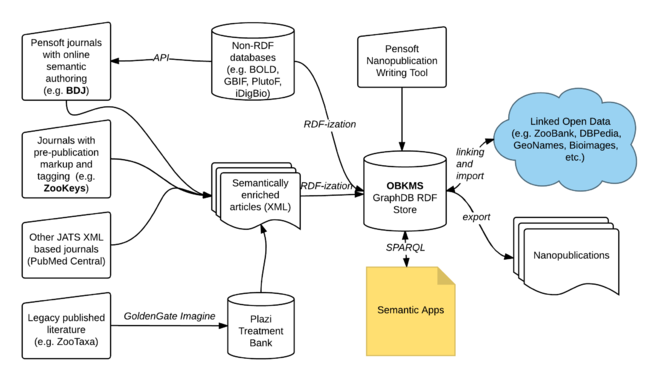
OpenBiodiv has several components:
- OpenBiodiv ontology: general data model allowing the extraction of biodiversity knowledge from taxonomic articles or from databases such as GBIF. The ontology (in preparation, Journal of Biomedical Semantics, available on GitHub) incorporates several pre-existing models: Darwin-SW (
Baskauf and Webb 2016 ), SPAR (Peroni 2014 ), Treatment Ontology, and several others. It defines classes, properties, and rules allowing to interlink these disparate ontologies and to create a LOD of biodiversity knowledge. New is the Taxonomic Name Usage class, accompanied by a Vocabulary of Taxonomic Statuses (created via an analysis of 4,000 Pensoft articles) allowing for the automated inference of the taxonomic status of Latinized scientific names. The ontology allows for multiple backbone taxonomies via the introduction of a Taxon Concept class (equivalent to DarwinCore Taxon) and Taxon Concept Labels as a subclass of biological name. - The Biodiversity Knowledge Graph - a LOD dataset of information extracted from taxonomic literature and databases. In practice, it has realized part of what has been proposed during pro-iBiosphere and later discussed by
Page (2016) . Its main resources are articles, sub-article componets (tables, figures, treatents, references), author names, institution names, geographical locations, biological names, taxon concepts, and occurrences. Authors have been disambiguated via their affiliation with the use of fuzzy-logic based on the GraphDB Lucene connector. The graph interlinks: (1) Prospectively published literature via Pensoft Publishers. (2) Legacy literature via Plazi. (3) Well-known resources such as geographical places or institutions via DBPedia. (4) GBIF's backbone taxonomy as a default but not preferential hierarchy of taxon concepts. (5) OpenBiodiv identifiers are matched to nomenclator identifiers (e.g. ZooBank) whenever possible. Names form two networks in the graph: (1) A directed-acyclical graph (DAG) of supercedence that can be followed to the corresponding sinks to infer the currently applicable scientific name for a given taxon. (2) A network of bi-directional relations indicating the relatedness of names. These names may be compared to the related names inferred on the basis of distributional semantics by the co-organizers of this workshop (Nguyen et al. 2017 ). - ropenbio: an R package for RDF*
3 -ization of biodiversity information resources according to the OpenBiodiv ontology. It will be submitted to the rOpenSci project. While many of its high-level functions are specific to OpenBiodiv, the low-level functions, and its RDF-ization framework can be used for any R-based RDF-ization effort. - OpenBiodiv.net: a front-end of the system allowing users to run low-level SPARQL queries as well to use an extensible set of semantic apps running on top of the Biodiversity Knowledge Graph.
Keywords
Linked Open Data, R package, RDF, SPARQL, Biodiversity Knowledge Graph, Semantic web, Semantic publishing, inference, Artificial Intelligence, Text Mining
Presenting author
Viktor Senderov and Teodor Georgiev
References
-
Darwin-SW: Darwin Core-based terms for expressing biodiversity data as RDF.Semantic Web7(6):629‑643. https://doi.org/10.3233/sw-150203
-
Constructing a biodiversity terminological inventory.PLOS ONE12(4):e0175277. https://doi.org/10.1371/journal.pone.0175277
-
Towards a biodiversity knowledge graph.Research Ideas and Outcomes2:e8767. https://doi.org/10.3897/rio.2.e8767
-
The Semantic Publishing and Referencing Ontologies.Law, Governance and Technology Series. https://doi.org/10.1007/978-3-319-04777-5_5
-
The Open Biodiversity Knowledge Management System in Scholarly Publishing.Research Ideas and Outcomes2:e7757. https://doi.org/10.3897/rio.2.e7757
LOD - Linked Open Data, the concept of interlinking data on the web introduced by Tim Berners-Lee, creator of the Web
SPARQL - Simple Protocol and Resource Description Framework Query Language
RDF - Resource Description Framework, a simple semantic format of knowledge representation inspired from linguistics